Papers & preprints
at Genentech (scientist)
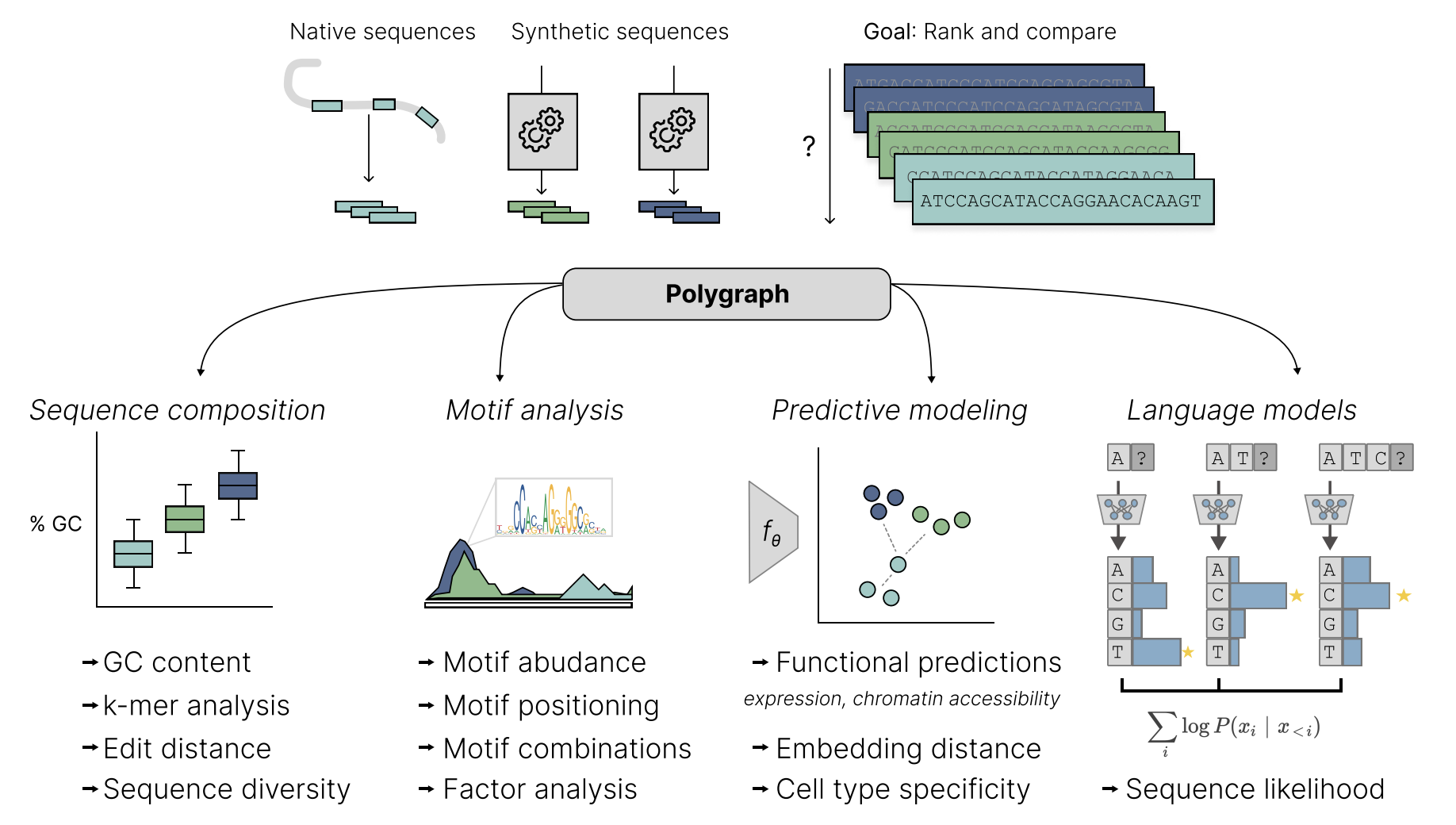
- Gupta, A.*, Lal, A.*, Gunsalus, L. M.*, Biancalani, T., & Eraslan, G. (2023). Polygraph: A Software Framework for the Systematic Assessment of Synthetic Regulatory DNA Elements. bioRxiv. Link to manuscript
at UCSF (PhD)
- Gunsalus, L. M. Sequence to structure to function: computational strategies for modeling the 3D genome. University of California, San Francisco, 2023. Link
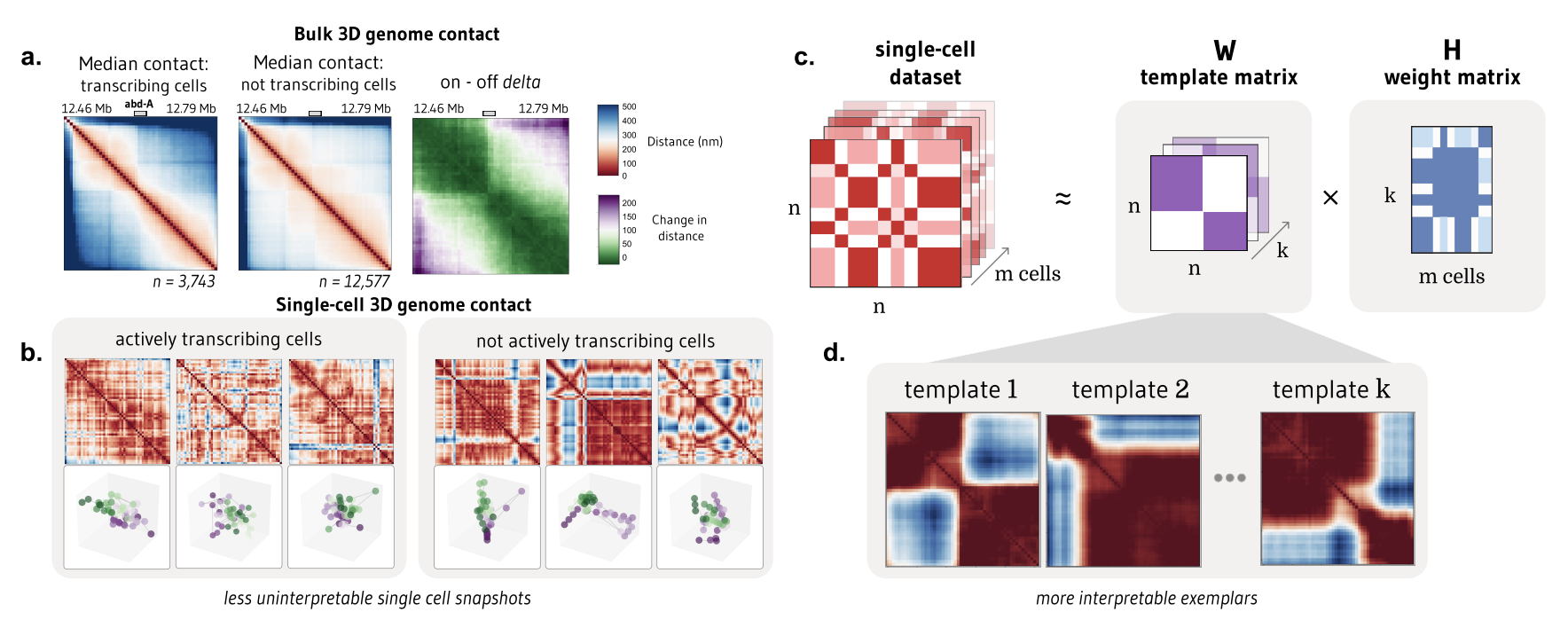
- Gunsalus, L. M., Michael J. Keiser, and Katherine S. Pollard. “ChromaFactor: deconvolution of single-molecule chromatin organization with non-negative matrix factorization.” bioRxiv (2023). Link to manuscript
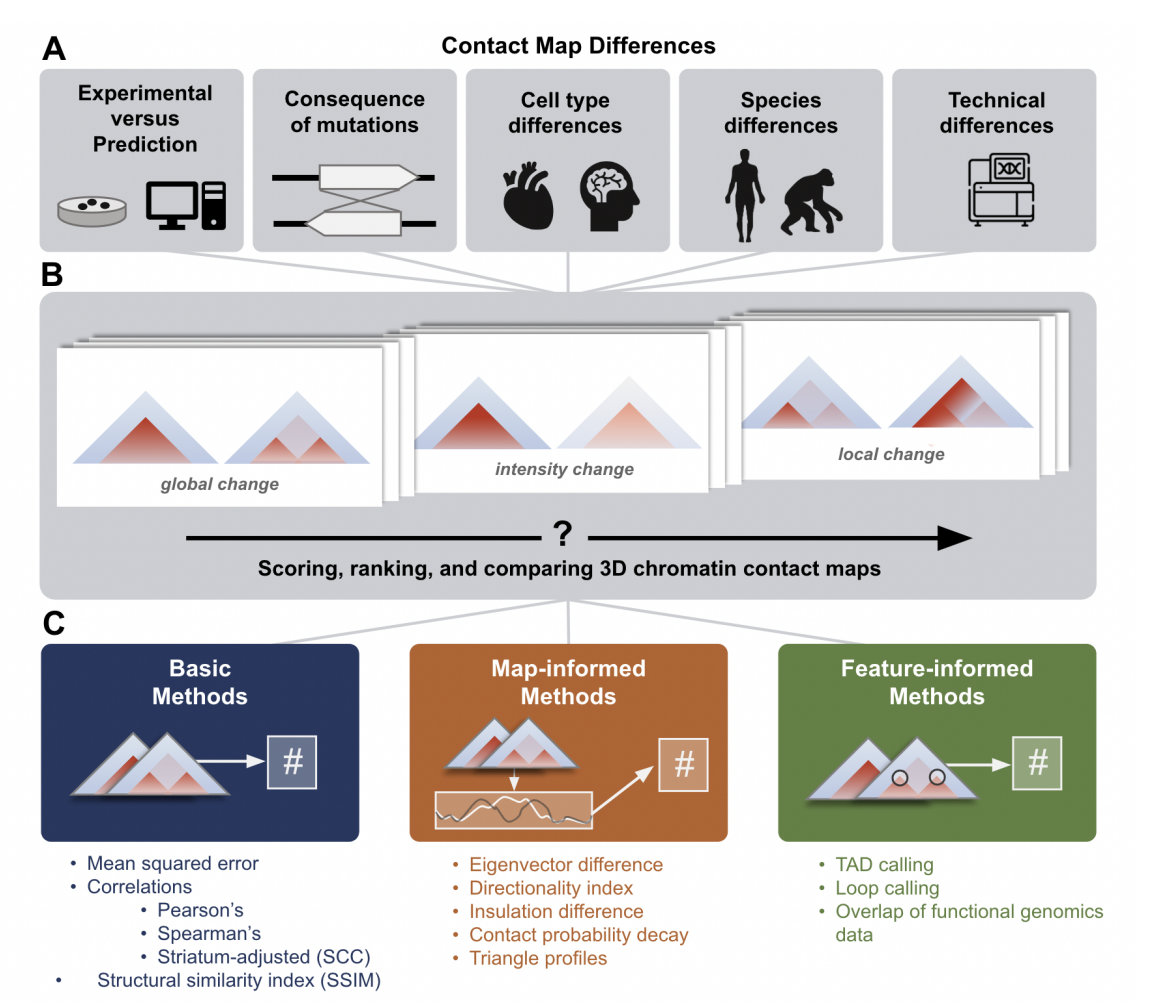
- Gunsalus, L. M.*, McArthur, E.*, Gjoni, K., Kuang, S., Pittman, M., Capra, J. A., & Pollard, K. S. (2023). Comparing chromatin contact maps at scale: methods and insights. bioRxiv. Link to manuscript
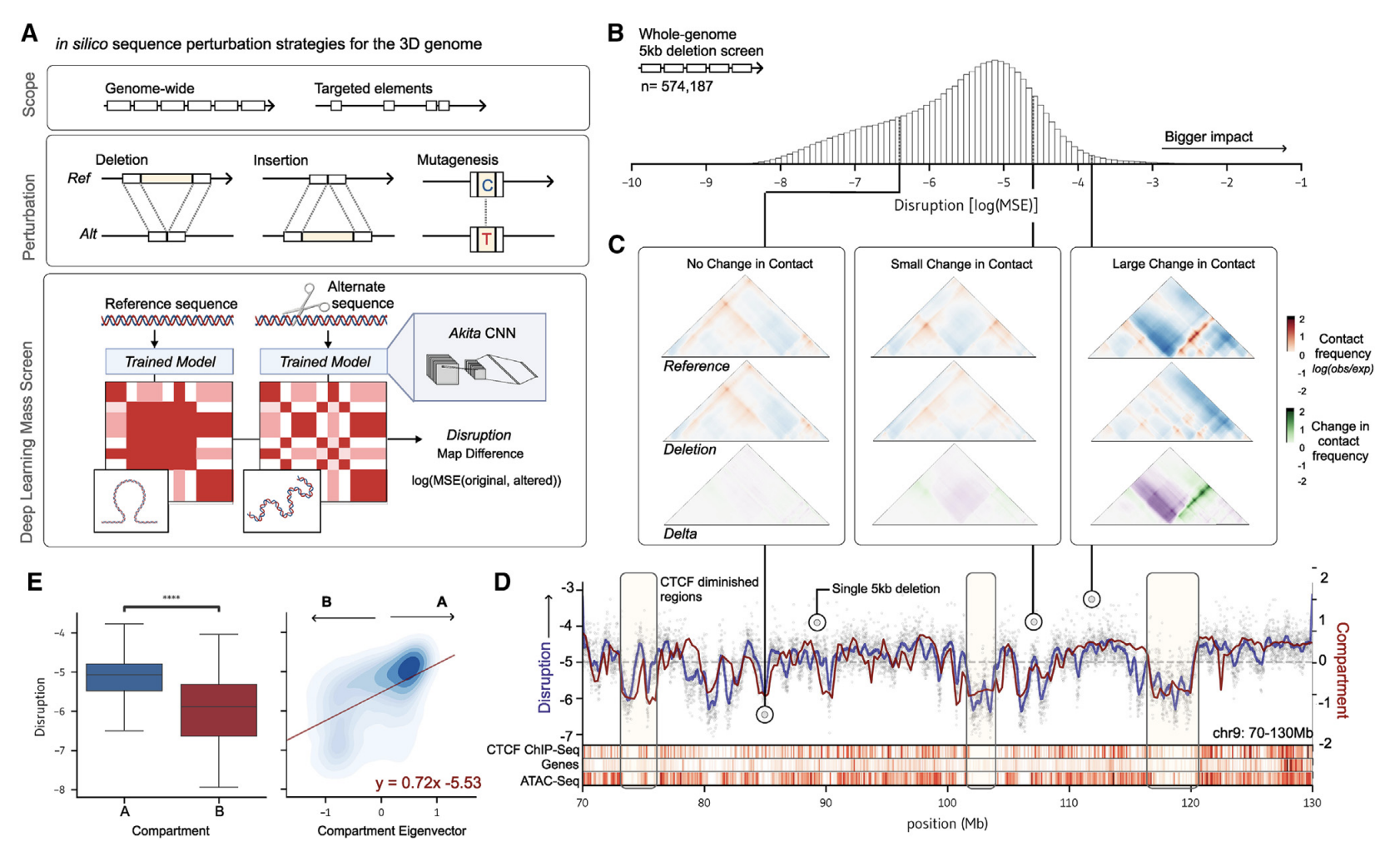
- Gunsalus, L. M., Keiser, M. J., & Pollard, K. S. (2023). In silico discovery of repetitive elements as key sequence determinants of 3D genome folding. Cell Genomics, 3(10). Link to paper
- Chuang, K. V., Gunsalus, L. M., & Keiser, M. J. (2020). Learning molecular representations for medicinal chemistry: miniperspective. Journal of Medicinal Chemistry, 63(16), 8705-8722. Link to paper
at Carnegie Mellon (undergrad)
- Ramamurthy, E., Welch, G., Cheng, J., Yuan, Y., Gunsalus, L., Bennett, D. A., … & Pfenning, A. R. (2023). Cell type-specific histone acetylation profiling of Alzheimer’s disease subjects and integration with genetics. Frontiers in Molecular Neuroscience, 15, 948456. Link to paper
* these authors contributed equally